
Those can help us use methods such as group_by more programmatically.ĭplyr uses non-standard evaluation (NSE) in all the important single table verbs: `filter()`, `mutate()`, `summarise()`, `arrange()`, `select()` and `group_by()`. Many dplyr verbs have an alternative version with an extra underline at the end. mutate( AGEGROUP = ifelse(RIDAGEYR %in% 0 : 18, "G0TO18", ifelse(RIDAGEYR %in% 19 : 35, "G19TO35", ifelse(RIDAGEYR %in% 36 : 65, "G36TO65", ifelse(RIDAGEYR %in% 66 : 79, "G66TO79", "G80"))))) %>% # NOTE : this would be a place to split the pipeline in 2 sections # Group by gender first, age second group_by(RIAGENDR, AGEGROUP) %>% # Summarize: count totals, summarise( Men = sum(RIAGENDR = 1), Women = sum(RIAGENDR = 2), MeanPFASR = formatC( mean(RATIO), format = "e", digits = 2), sdPFASR = formatC( sd(RATIO), format = "e", digits = 2), MeanTChol = mean(LBXTC), sdTChol = sd(LBXTC), minTChol = min(LBXTC), maxTChol = max(LBXTC) ) `summarise()` has grouped output by 'RIAGENDR'.Function with quosure -Screenshot by the author The pipeline below is annotated to specify the function of each line and the results are saved within a user-defined R object Example2.Įxample2 % # select columns select(SEQN, RIAGENDR, RIDAGEYR, LBXMFOS, URXUCR, LBXTC ) %>% # fitler all rows to remove NAs drop_na() %>% # Creatinine adjustment mutate ( RATIO = (LBXMFOS /URXUCR) * 10 ^- 4) %>% # categorize ages in 5 groups: # Children: G0TO18, younger adults: G19TO35, # and older adults: G36TO65, seniors: G66TO79, # and 80 and older: G80. (Try without and see the difference! - this option was suggested on Stack Overflow.) We’ll have to use the appropriate columns of data, compute the creatinine adjustment and summarize these values by age group for men and women.Ī new base R function is introduced here formatC() which will force the numbers to be printed in scientific exponent for better clarity. (For a refresher on ifelse() see section 10.4.1.) Nesting the ifelse() function within itself allows making multiple choices.
One of the key word is “age group” as in the NHANES data age is a single integer number with a range from 1 to 80 in RIDAGEYR column and therefore it is “up to us” to create the age groups! This can be done rather easily within a combination of mutate() and with nested ifelse() statements within. “ Base on gender and age group, what is the mean and standard deviation of PFAS compounds, as well as the mean, standard deviation, minimum, and maximum values of total cholesterol in men and women?” Let’s make things a bit more interesting with the question: We’ll place the results in a user-defined R object so that we can reuse it in the next section without re-writing the complete pipeline. For each marital status code, we’ll then count how many women and men are in each category that we’ll report in columns Mnum and Wnum (the sum of these 2 on each line should be equal to the reported Counts column.)įor simplicity with income and education we’ll simply compute the mean of the codes, which should still give us an indication of the level of income and education for each category of marital status. We want to get some information by group, in this case we’ll group by marital status and then count the total number of observations for each case and we’ll store this in column Counts. DMDEDUC2 - Education level - Adults 20+.RIAGENDR: 1-male, 2-female (none missing).The following data can be found in the DEMO_I demographic data file: I The Story of Vector V: an R markdown example.D.5 Download, save XPT files to hard drive.
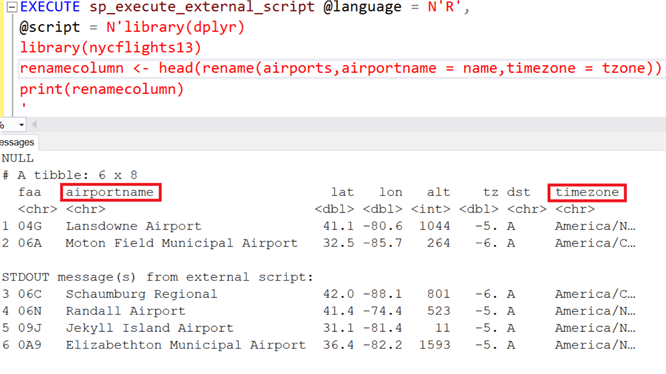
#Dplyr rename download
#Dplyr rename code
D.1 Download into R object with NHANES code.14.4 Preamble, Preface and Introduction.
#Dplyr rename how to


 0 kommentar(er)
0 kommentar(er)
